BESSY II 202406 Flow no flow Pocillopora spp
BESSY II 2024. Pocillopora spp. Flow / No flow
Resume of the project
In the island of Mo’orea located in French Polynesia, Pocillopora spp. are present and conciderate as dominante species around the island (back reef - fore reef) following many event as Crown of Throne, bleaching event (Gonzalo Pérez-Rosales et al. 2021). Pocillopora spp. show lot of plasticity depending of the depth, light and courant (Johnston et al. 2017). Identified Pocillopora spp. at species level are complicated only by morphology. Molecular work need to be done to detect and identified the species level using mtORF (Johnston et al. 2022). In this experiment we identify 4 differents species P.verrucosa (Haplotype 3), P. meandrina (Haplotype 1a - 8a), P. tuahiniensis (Haplotype 10)(new specie describe by Johnston and Burgess 2023), P. grandis (Haplotype 1a) sampled at 2 differents sites conditions; Haapiti Flow site (W 149.883782438236 - S 17.5650093638768)and Manava No flow site (W 149.807646130636 - S 17.4756927846393). Our goal it’s to identified the difference and the plasticity the coral make between a site with a hight flow regime compare to a low flow regime condition. For doing that we run tomography scanning (Scucchia et al. 2023) on our samples at the BeamLine of BESSY II in Berlin.
Idea
Depending of the environmental condition, Pocillopora spp. will don’t have the same skeleton structure interspecific and intraspecific. Our first try will be on aldult colonie and we will keep pushing on this aspect for the recruit.
History of the adults samples
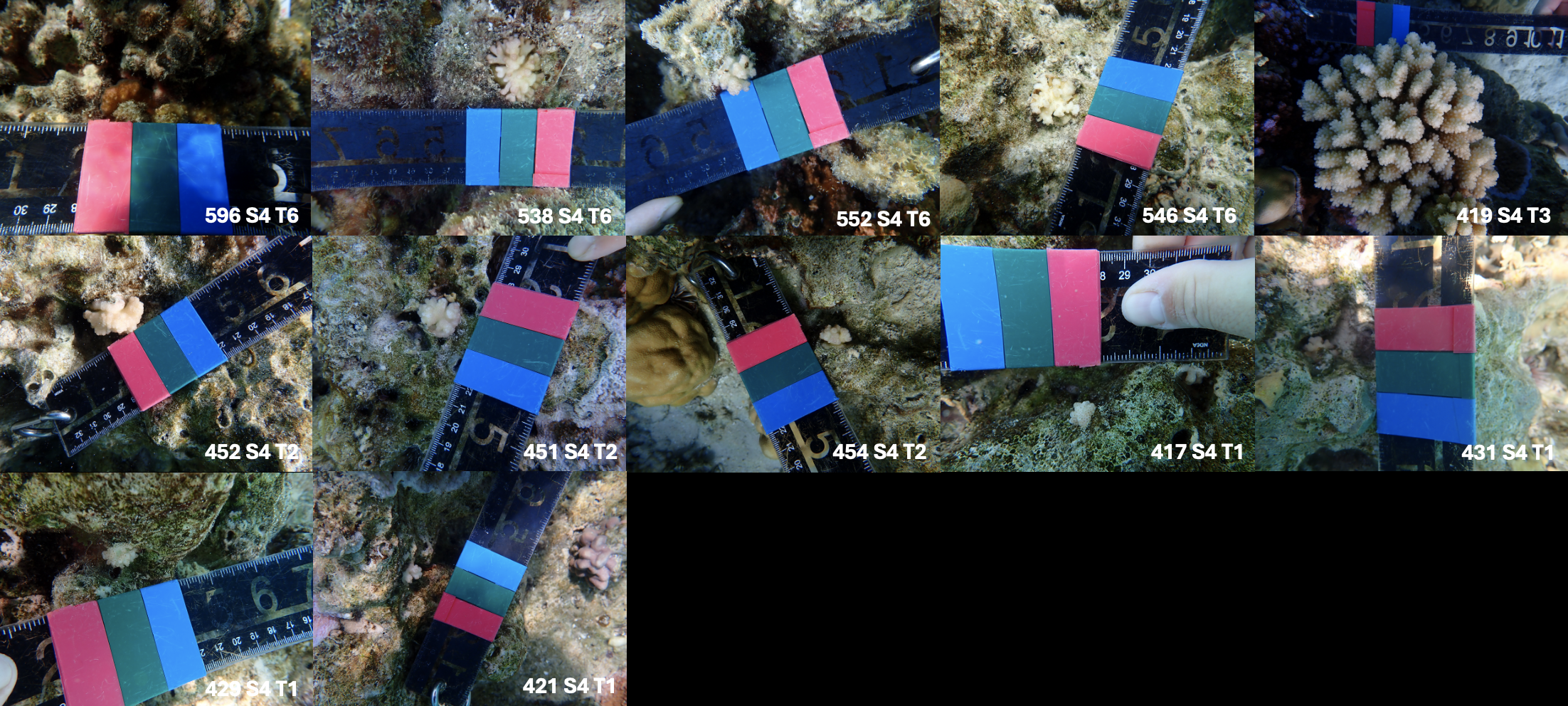
Colonies from the field

History of the recruits samples
Colonies from the field
In order to prepare a new approach to the question with new life stages, we used the same approach as for adults. However, among the recruits chosen, only one site is present, due to a lack of time to obtain sample IDs. This approach will serve as a test to understand the potential for comparison between life stages and species for a future Beamtime.
Samples preparation adults
All samples were prepared in advance following the protocol SEM preparation. Before each scan, the coral fragments were cut with a pair of wire cutters to take only the top of the fragments (see picture bellow).

Samples preparation recruits
All samples were prepared in advance following the protocol SEM preparation. All recruits were cut or positioned to be able to see the entire fragment. Each fragment was scanned twice, starting from the top tips and going down 3 mm to overlap the scans for easier reconstruction.
Sample scan
Adults flow no flow for both site (Haapiti - Manava)
| Date | Time | Sample name | File name | Number of scan | Resolution | Notes |
|---|---|---|---|---|---|---|
| 20240629 | 21:10 | PT013 | 2 | |||
| 20240629 | 21:20 | PT040 | PT410 | 2 | ||
| 20240629 | 21:43 | PG010 | PG110 | 1 | Tiny sample, no need 2 scan to cover the entire sample | |
| 20240629 | 21:57 | PM029 | PM29 | 2 | ||
| 20240629 | 22:27 | PV002 | PV02 | 2 | ||
| 20240629 | 22:52 | PT042 | PT12 | 1 | Only 1 scan | |
| 20240629 | 23:08 | PV035 | PV35 | 1 | Only 1 scan | |
| 20240629 | 23:22 | PT015 | PT15 | 1 | Only 1 scan | |
| 20240629 | 23:42 | PG030 | PG310 | 2 | ||
| 20240630 | 0:43 | PM021 | PM21 | 2 | ||
| 20240630 | 1:10 | POC NO FLOW | POCC NO FLOW | 2 | ||
| 20240630 | 1:30 | POC FLOW | POCC FLOW | 2 | ||
| 20240630 | 2:00 | PG034 | PG34 | 2 | ||
| 20240630 | 2:32 | PG011 | PG11 | 2 | ||
| 20240630 | 3:05 | PV036 | PV36 | 2 | ||
| 20240630 | 3:32 | PV001 | PV01 | 2 | ||
| 20240630 | 3:57 | PM019 | PM19 | 2 | ||
| 20240630 | 4:20 | C2 | PDAM_C2 | 1 | Problem during scan, catch lot of paper and not the sample | |
| 20240630 | 4:35 | M3 | PDAM_M3 | 1 | Problem during scan, catch lot of paper and not the sample | |
| 20240630 | 4:49 | H3 | PDAM_H3 | 1 | Problem during scan, catch lot of paper and not the sample | |
| 20240630 | 5:03 | C1 | PDAM_C1 | 1 | Problem during scan, catch lot of paper and not the sample | |
| 20240630 | 5:30 | 419 | POCC13 | 2 | ||
| 20240630 | 5:45 | 596 | POCC13 | 2 | ||
| 20240630 | 6:25 | FUNGIA | FUN1 | 2 |
Sample scan
Recruits from Manava site (no flow)
| Date | Species | Sample number | Sample file name | Sample file name rest | Number of scan | Notes |
|---|---|---|---|---|---|---|
| 20240630 | Haplotype 1a | 596 | POCC | 1 | 2 | |
| 20240630 | P.meandrina | 452 | POCC | 2 | 2 | |
| 20240630 | P.effusa | 417 | POCC | 3 | 2 | |
| 20240630 | Haplotype 1a | 431 | POCC | 4 | 2 | |
| 20240630 | P.meandrina | 538 | POCC | 5 | 2 | |
| 20240630 | Haplotype 1a | 451 | POCC | 6 | 2 | the second scan is 4mm bellow |
| 20240630 | Haplotype 1a | 429 | POCC | 7 | 2 | |
| 20240630 | P.tuahiniensis | 421 | POCC | 8 | 2 | |
| 20240630 | P.acuta | 454 | POCC | 9 | 2 | |
| 20240630 | P.tuahiniensis | 552 | POCC | 10 | 2 | |
| 20240630 | P.acuta | 546 | POCC | 11 | 2 | |
| 20240630 | P.effusa | 419 | POCC | 13 | 2 |
Sample scan parameters
All samples adults/recruits
| Folder | File name | Sample file name | Energy (eV) | Resolution | Exposition time |
|---|---|---|---|---|---|
| PV3 | _00340 | PV36 | 24500 | 2X | 150ms |
| PV3 | _00338 | PV36 | 24500 | 2X | 150ms |
| PV3 | _00310 | PV35 | 24500 | 2X | 150ms |
| PV3 | _00213 | PV36 | 24500 | 10X | 750ms |
| PV3 | _00212 | PV36 | 24500 | 10X | 100ms |
| PV3 | _00211 | PV36 | 24500 | 10X | 100ms |
| PV3 | _00210 | PV36 | 24500 | 10X | 600ms |
| PV3 | _00209 | PV36 | 24500 | 10X | 450ms |
| PV3 | _00208 | PV36 | 24500 | 10X | 150ms |
| PV3 | _00207 | PV36 | 24500 | 10X | 150ms |
| PV3 | _00206 | PV36 | 24500 | 10X | 150ms |
| PV3 | _00204 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00203 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00202 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00201 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00200 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00199 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00198 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00197 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00196 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00191 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00190 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00189 | PV36 | 24523 | 10X | 150ms |
| PV3 | _00188 | PV36 | 24523 | 10X | 150ms |
| PV0 | _00344 | PV01 | 24500 | 2X | 150ms |
| PV0 | _00342 | PV01 | 24500 | 2X | 150ms |
| PV0 | _00306 | PV02 | 24500 | 2X | 150ms |
| PV0 | _00304 | PV02 | 24500 | 2X | 150ms |
| PV0 | _00227 | PV01 | 17000 | 10X | 750ms |
| PV0 | _00224 | PV01 | 24500 | 10X | 750ms |
| PV0 | _00222 | PV01 | 16000 | 10X | 750ms |
| PV0 | _00220 | PV01 | 24500 | 10X | 750ms |
| PT4 | _00296 | PT410 | 24500 | 2X | 150ms |
| PT4 | _00295 | PT410 | 24500 | 2X | 150ms |
| PT4 | _00293 | PT410 | 24500 | 2X | 150ms |
| PT1 | _00312 | PT15 | 24500 | 2X | 150ms |
| PT1 | _00308 | PT12 | 24500 | 2X | 150ms |
| PT1 | _00291 | PT13 | 24500 | 2X | 150ms |
| PT1 | _00289 | PT13 | 24500 | 2X | 150ms |
| Pocc | _00396 | Pocc2 | 24500 | 2X | 150ms |
| Pocc | _00395 | Pocc2 | 24500 | 2X | 150ms |
| Pocc | _00394 | Pocc3 | 24500 | 2X | 150ms |
| Pocc | _00392 | Pocc3 | 24500 | 2X | 150ms |
| Pocc | _00390 | Pocc4 | 24500 | 2X | 150ms |
| Pocc | _00388 | Pocc4 | 24500 | 2X | 150ms |
| Pocc | _00386 | Pocc6 | 24500 | 2X | 150ms |
| Pocc | _00385 | Pocc6 | 24500 | 2X | 150ms |
| Pocc | _00383 | Pocc7 | 24500 | 2X | 150ms |
| Pocc | _00381 | Pocc7 | 24500 | 2X | 150ms |
| Pocc | _00379 | Pocc8 | 24500 | 2X | 150ms |
| Pocc | _00378 | Pocc9 | 24500 | 2X | 150ms |
| Pocc | _00376 | Pocc9 | 24500 | 2X | 150ms |
| Pocc | _00374 | Pocc10 | 24500 | 2X | 150ms |
| Pocc | _00372 | Pocc10 | 24500 | 2X | 150ms |
| Pocc | _00370 | Pocc11 | 24500 | 2X | 150ms |
| Pocc | _00368 | Pocc11 | 24500 | 2X | 150ms |
| Pocc | _00365 | Pocc5 | 24500 | 2X | 150ms |
| Pocc | _00363 | Pocc5 | 24500 | 2X | 150ms |
| Pocc | _00360 | Pocc1 | 24500 | 2X | 150ms |
| Pocc | _00358 | Pocc1 | 24500 | 2X | 150ms |
| Pocc | _00356 | Pocc13 | 24500 | 2X | 150ms |
| Pocc | _00328 | Pocc flow | 24500 | 2X | 150ms |
| Pocc | _00326 | Pocc flow | 24500 | 2X | 150ms |
| Pocc | _00324 | Pocc no flow | 24500 | 2X | 150ms |
| Pocc | _00322 | Pocc no flow | 24500 | 2X | 150ms |
| PM4 | _00252 | PM25 | 17000 | 10X | 450ms |
| PM4 | _00250 | PM25 | 24500 | 10X | 450ms |
| PM2 | _00320 | PM21 | 24500 | 2X | 150ms |
| PM2 | _00318 | PM21 | 24500 | 2X | 150ms |
| PM2 | _00302 | PM29 | 24500 | 2X | 150ms |
| PM2 | _00300 | PM29 | 24500 | 2X | 150ms |
| PM2 | _00287 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00286 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00284 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00282 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00281 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00279 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00277 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00276 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00274 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00273 | PM25 | 24500 | 2X | 150ms |
| PM2 | _00266 | PM25 | 17000 | 10X | 450ms |
| PM2 | _00264 | PM25 | 24500 | 10X | 450ms |
| PM2 | _00263 | PM25 | 24500 | 10X | 450ms |
| PM1 | _00348 | PM19 | 24500 | 2X | 150ms |
| PM1 | _00346 | PM19 | 24500 | 2X | 150ms |
| PM1 | _00243 | PM19 | 24500 | 10X | 450ms |
| PM1 | _00241 | PM19 | 17000 | 10X | 450ms |
| PM1 | _00238 | PM19 | 24500 | 10X | 450ms |
| PG3 | _00332 | PG34 | 24500 | 2X | 150ms |
| PG3 | _00330 | PG34 | 24500 | 2X | 150ms |
| PG3 | _00316 | PG310 | 24500 | 2X | 150ms |
| PG3 | _00314 | PG310 | 24500 | 2X | 150ms |
| PG3 | _00249 | PG34 | 24500 | 10X | 450ms |
| PG3 | _00248 | PG34 | 24500 | 10X | 450ms |
| PG3 | _00246 | PG34 | 17000 | 10X | 450ms |
| PG3 | _00244 | PG34 | 24500 | 10X | 450ms |
| PG1 | _00336 | PG11 | 24500 | 2X | 150ms |
| PG1 | _00334 | PG11 | 24500 | 2X | 150ms |
| PG1 | _00298 | PG110 | 24500 | 2X | 150ms |
| PG1 | _00237 | PG11 | 24500 | 10X | 450ms |
| PG1 | _00234 | PG11 | 17000 | 10X | 450ms |
| PG1 | _00232 | PG11 | 24500 | 10X | 450ms |
| PG1 | _00231 | PG11 | 24500 | 10X | 450ms |
All the data is available on GitHub here